Table Of Contents
Previous topic
Next topic
Quick search
Enter search terms or a module, class or function name.
The package data handles the interaction between the ProbReM instance and the relational database.
The core idea of the data interface is to separate the PRM model from the relational data soucre. The module datainterface contains a collection of methods to access the relational data that are used by different algorithms (e.g. CPD learners, inference methods, EM algorithm). Another advantage of this approach is that the data is not required to be in a fixed database format.
List that contains all DataSetInterface instances that connect to the data
An instance of the class connects a PRM with the relational data base that it models. This is an abstract class, sublclasses have to implement the required methdos for different database systems. E.g. data.sqliteinterface.SQLiteDI is for SQLite.
There data is queried by the parameter learning algorithm, e.g. learners.cpdlearners, as well as by the inference methods; the inference engine is unrolling the Ground Bayesian Network.
Returns a dictionary mapping a crossvalidation test set with the corresponding training set { datasetinstance : [datasetinstance1,datasetinstance2,....]}. Every DataSetInterface in datainterface.DSI is a key in datainterface.trainingSets, the value is a list of all other DataSetInterface instances in datainterface.DSI
A method that allows us to configure a dataset interface based on information from the instantiated PRM.
Creates a connection to a database. There are possibly multiple dataset connections to do crossvalidation.
| Parameters: |
|
|---|---|
| Returns: | A DataSetInterface instance |
The type of dataset used, e.g. crossvalidation or testtraining
The name of the data interface
Dictionary that maps a crossvalidation test set (one DataSetInterface in datainterface.DSI) with the corresponding training set (all other DataSetInterfaces in datainterface.DSI)
The class DataSetInterface specifies a set of methods that are need to be implemented by a database specific data interface. Currently the framework supports only SQLite which is implemented in data.sqliteinterface.SQLiteDI. Implementing support for other SQL based database systems, e.g. MySQL, should be straight forward. Support for other database systems would require more effort.
A subclass of DataSetInterface that links a PRM to the SQLite database that it models.
If a probabilistic dependency between an attribute and a parent attribute is of type m:n or 1:n, some sort of aggregation has to be performed. A VIEW is created that performs the necessary aggregation and enables the datainterface to query the already aggregated values in one query. The name of the view is dep.name and it can be used when learning local distributions or unrolling a ground bayes net.
When there are multiple dependencies from and to the same erClass, then we could create only one view. Instead we create a view for each dependency independently which makes it easier but redundant
Note
VIEWS are implemented but not used. In practice their performance proved to be worse than working on the data directly.
SQLite cursor that will execute SQL commands and contain the result set
All attribute objects of the attribute attr are queried. The result set will consist of rows in the following format:
| Parameters: | attr – Attribute |
|---|
Given a set of children objects obj for a given attribute attr, we are loading the set of parents (for all depenencies that attr is a child of). This method is not used because it performs poor compared to loadDependencyChildrenObjects() and loadDependencyParentObjects().
We pass an attribute, and constructe a query such that the result set self.cur contains all the data to learn the local distribution of that attribute. The Count in the name indicates that the query is constructed such that the compuation is done on the SQL side, e.g. the number of occurences for each possible parent assignment is counted using COUNT. This function is used by learners.cpdlearners.CPDTabularLearner.learnCPDsCount().
| Parameters: | attribute – Subclass of prm.attribute.Attribute |
|---|
Given a set of parent attribute objects gbnVertices for a given dependency dep, we are loading the set of children.
The result set will consist of rows in the following format:
| Parameters: |
|
|---|
Given a set of children attribute objects gbnVertices for a given dependency dep, we are loading the set of parents.
The result set will consist of rows in the following format:
| Parameters: |
|
|---|
In the case of reference uncertainty, the exist attributes have a set of parents that need to be included in the ground Bayesian network. The SQL query needed is constructed in this method, the resultset will be of the following format. The k-entity references the entity on the k side of the n:k relationship (i.e. Professor in the student/prof example from Pasula). The primary key of the k-entity is used as identifier.
| Parameters: |
|
|---|
We pass an attribute, and constructe a query such that the result set self.cur contains all the data to learn the local distribution of that attribute. The Full in the name indicates that we don’t COUNT all occurences in the query but in the learner instead. The Agg indicates that if the attribute has multiple parents for one attribute object, we use VIEWS to aggregate the data using SQL.
In practice this has proven to not be a good way to do things. First, it is much slower than the other methods. Second, by aggregating the values before counting them the number of occurences per parent assignment is much smaller. On the other hand, by not aggregating them we introduce a bias in which attribute objects with many parent attribute objects for on parent attribute are weighted more. Thus this methods is currently not used.
This method is currently only used to compute the log likelihood of the model given the data using learners.cpdlearners.CPDTabularLearner.loglikelihood().
| Parameters: | attribute – Subclass of prm.attribute.Attribute |
|---|
We pass an attribute, and constructe a query such that the result set self.cur contains all the data to learn the local distribution of that attribute. The Full in the name indicates that we don’t COUNT all occurences in the query but in the learner instead. This method is used by learners.cpdlearners.CPDTabularLearner.learnCPDsFull().
| Parameters: | attribute – Subclass of prm.attribute.Attribute |
|---|
When unrolling a Ground Bayes Net the inference engine inference.engine processes a set of event and evidence variables that are of type inference.query.Qvariable. The method self.loadObjects() executes a SQL query that returns the set of all attribute objects that satisfy the constraints of qvar
The result set will have the following structure : [attribute, pk1 , pk2, ....], e.g.
- If qvar.erClass is User : [User.gender, User.user_id]
- If qvar.erClass is rates : [rates.rating, rates.user_id,rates.item_id]
| Parameters: | qvar – inference.query.Qvariable |
|---|
Path to SQLite DB file
We return the the cursor which is an iterable result set of the executed query (after executing a loadXXX() method). The result set can then be iterated like this in the caller method:
for currentRow in dsi.resultSet():
do something with `currentRow`
After executing a loadXXX() method, the cursor self.cur contains the result set for a specific SQL query. This method returns the next row in the result set, which allows a caller, e.g. learners.cpdlearners.CPDTabularLearner.learnCPDsFull() or inference.engine.unrollGBN(), to iterate over all rows without knowledge about the data interface.
Aggregation becomes necessary if an attribute object has multiple parent attribute objects for one single dependency, e.g. in the case of a 1:n or m:n relationship. As the conditional probability distribution (CPD ) of that attribute class allows only one parent value for this dependency, the values of these parent attribute objects must be aggregated. Any function 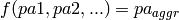 where
where 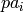 are the parent values and
are the parent values and 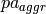 is a single value in the domain of the parent attribute class.
is a single value in the domain of the parent attribute class.
Aggregation can be performed on different levels, e.g. on the database level using SQL keywords or during runtime by aggregating values in the Ground Bayesian Network. The .aggregation module provides this functionality for a set of aggregation functions. When instantiating an aggregator type for a dependency, e.g. the MAX aggregator during runtime:
import data.aggregation
aggr = data.aggregation.aggregators['MAX']['runtime']
returns the method data.aggregation.runtime_avg(). Or
aggr = data.aggregation.aggregators['MAX']['SQLite']
simply returns the SQL keyword AVG.
SQLite keyword for maximum, MIN
Dictionary of supported aggregation types.
- AVG : ‘SQLite’,’runtime’ supported
- MAX : ‘SQLite’,’runtime’ supported
- MIN : ‘SQLite’,’runtime’ supported
- MODE : only ‘runtime’ supported
All variables are discrete, therefore we round to the nearest integer.
| Parameters: | values – List of attribute object values |
|---|---|
| Returns: | Average of values |
Compute MAX of all values.
| Parameters: | values – List of attribute object values |
|---|---|
| Returns: | Maximum of values |
A module containing utils for handling data.
Discretizer can be used to discretize continuous data. self.bins is a list of values starting with the smallest possible value and ending with the largest possible value. All intervals in between are bins, e.g. when discretize(self,value) is called the index of the bin that value falls in is returned.