Recent Research Highlights
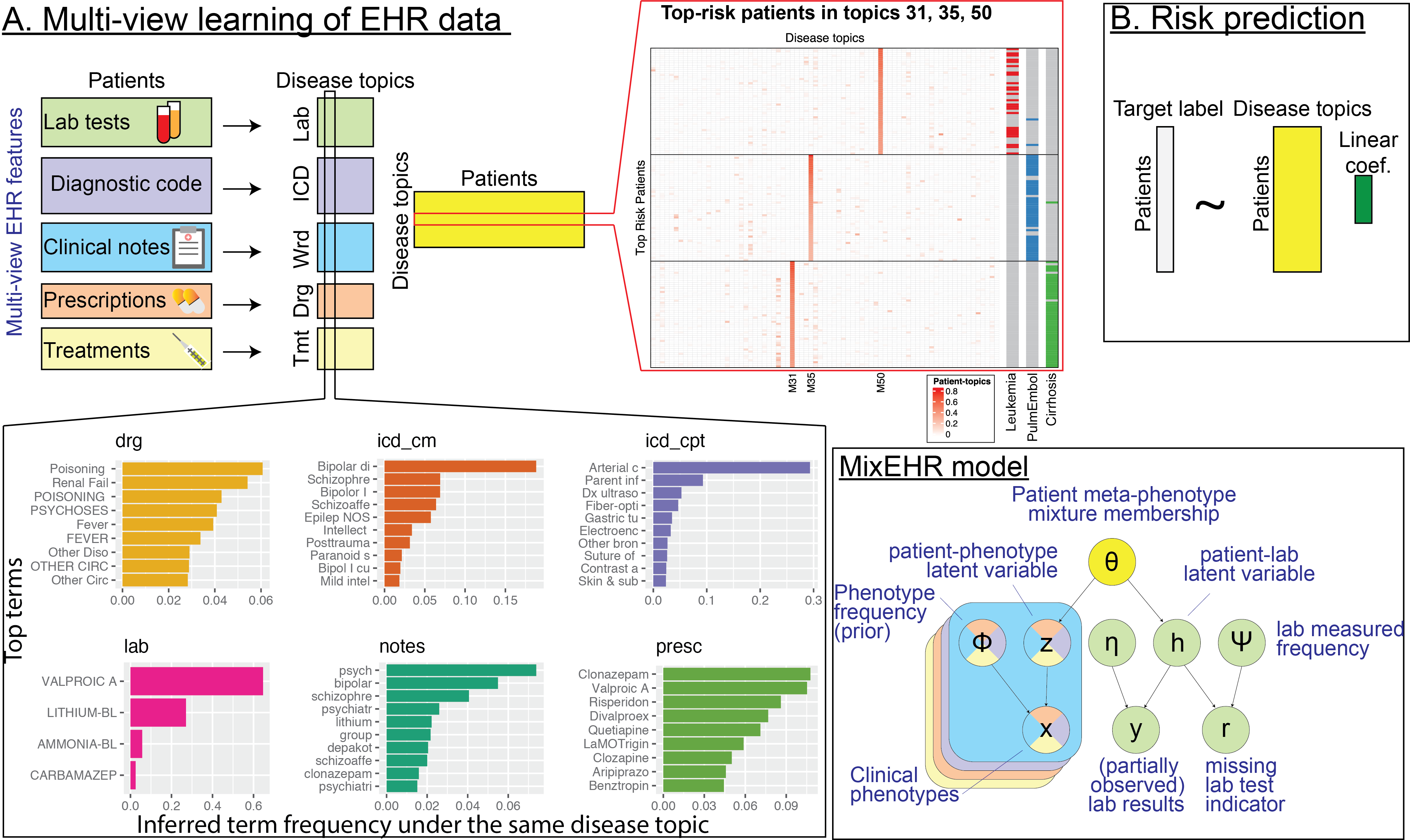
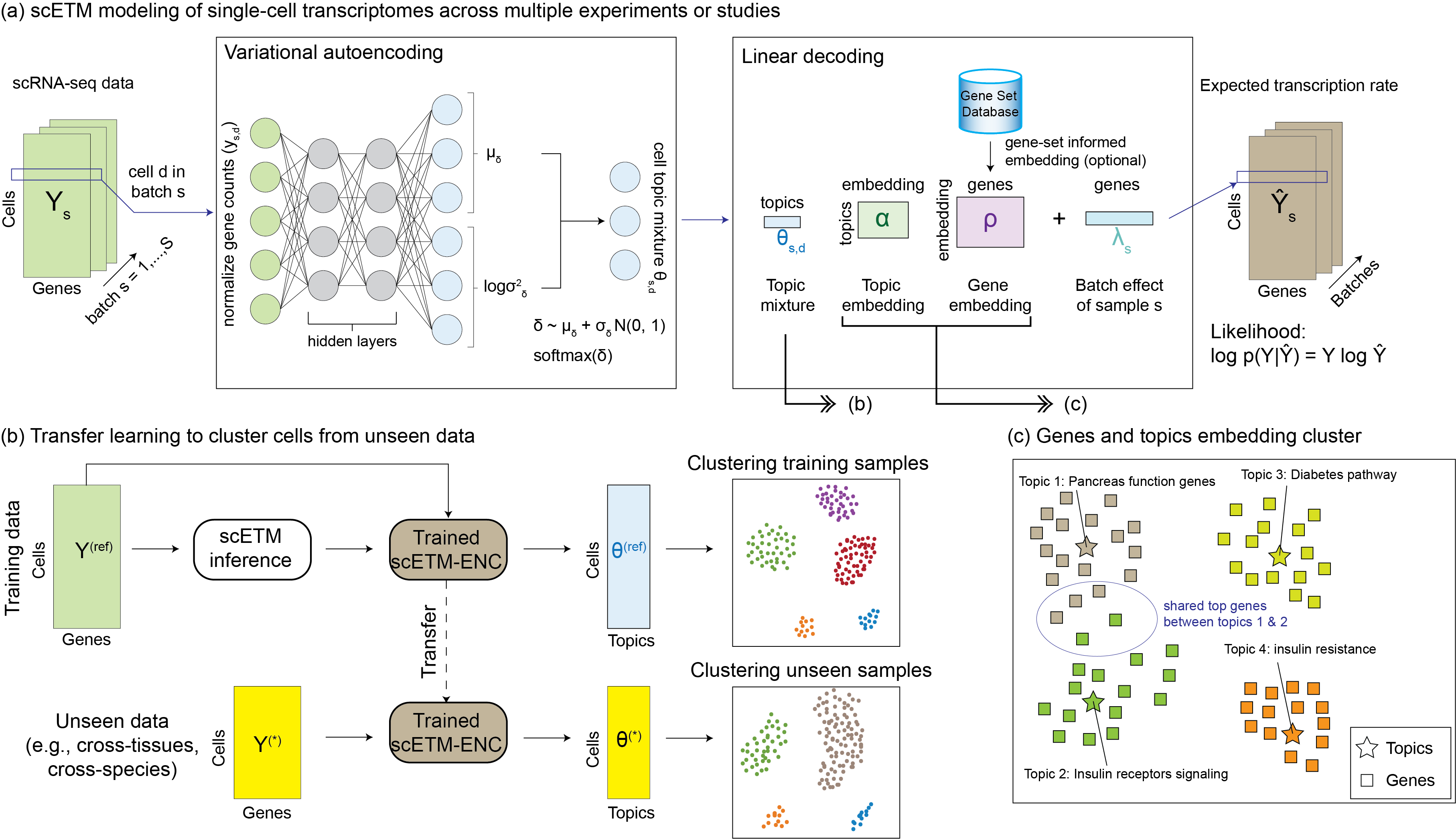
Multimodal EHR integration
Inferring multimodal latent topics from electronic health records. Nature Communications 2020
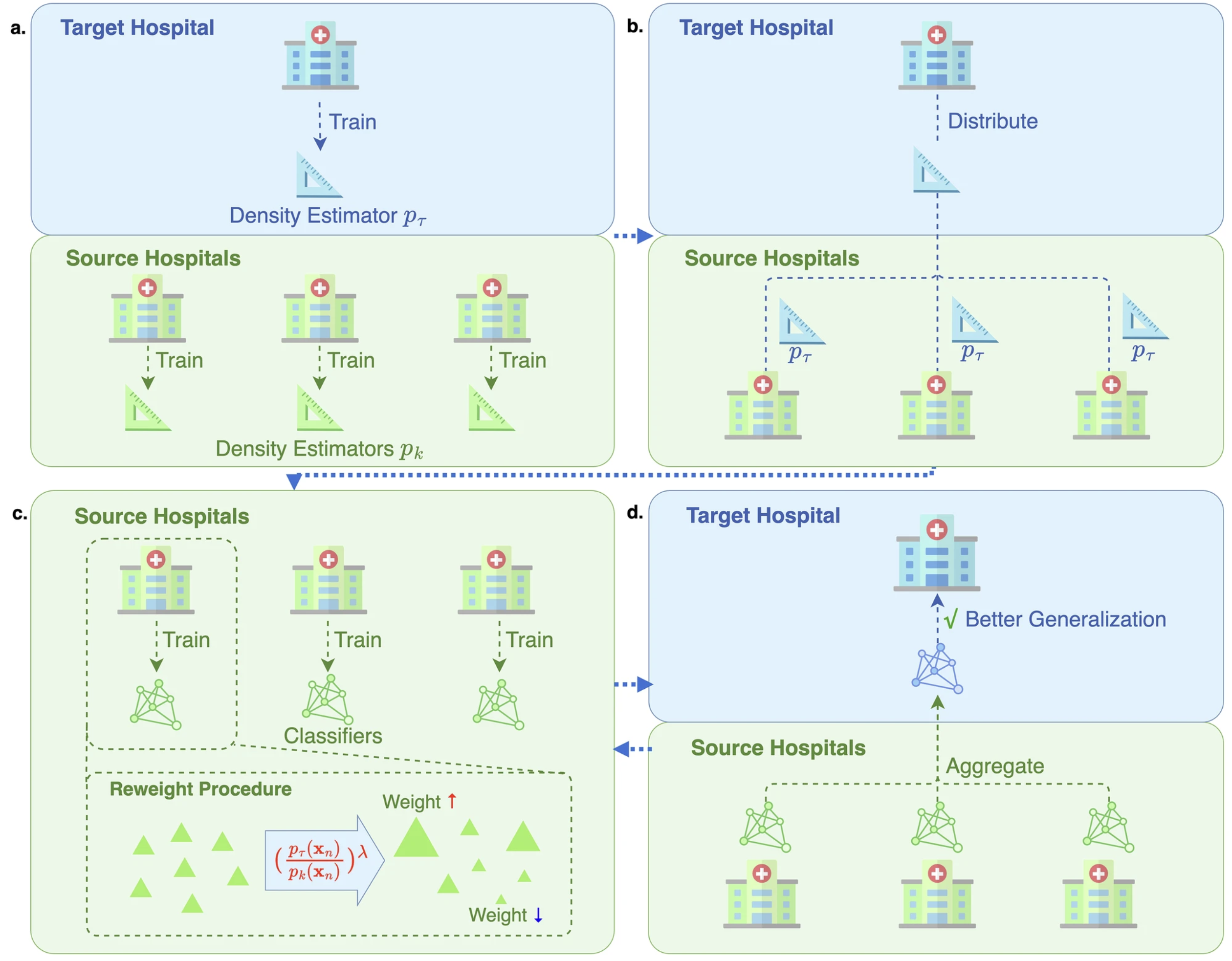
Federated learning in EHR
FedWeight: mitigating covariate shift of federated learning on EHR data through patients re-weighting. npj Digital Medicine 2025
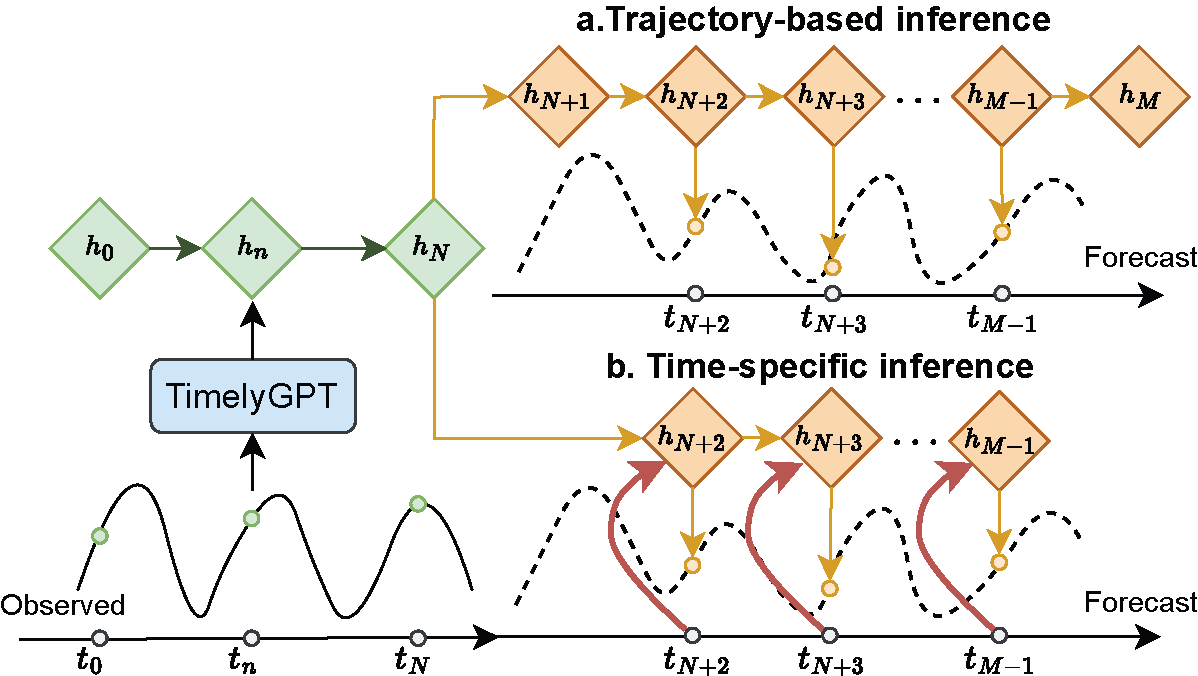
Time-series health forecasting
TimelyGPT: Extrapolatable Transformer Pre-training for Long-term Time-Series Forecasting in Healthcare. ACM-BCB 2024
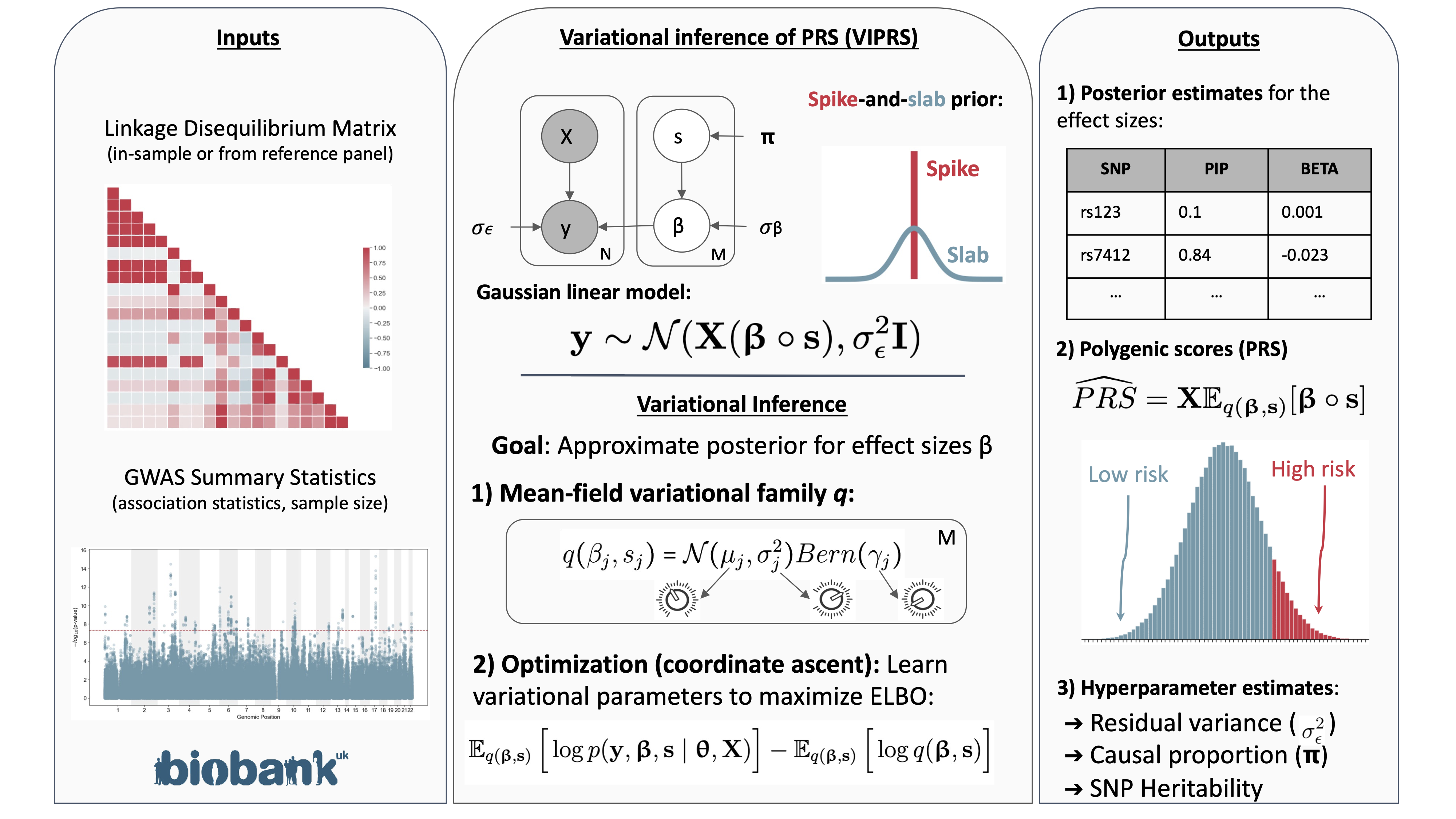
Polygenic risk score inference
Fast and Scalable Polygenic Risk Modeling with Variational Inference. American Journal of Human Genetics 2023
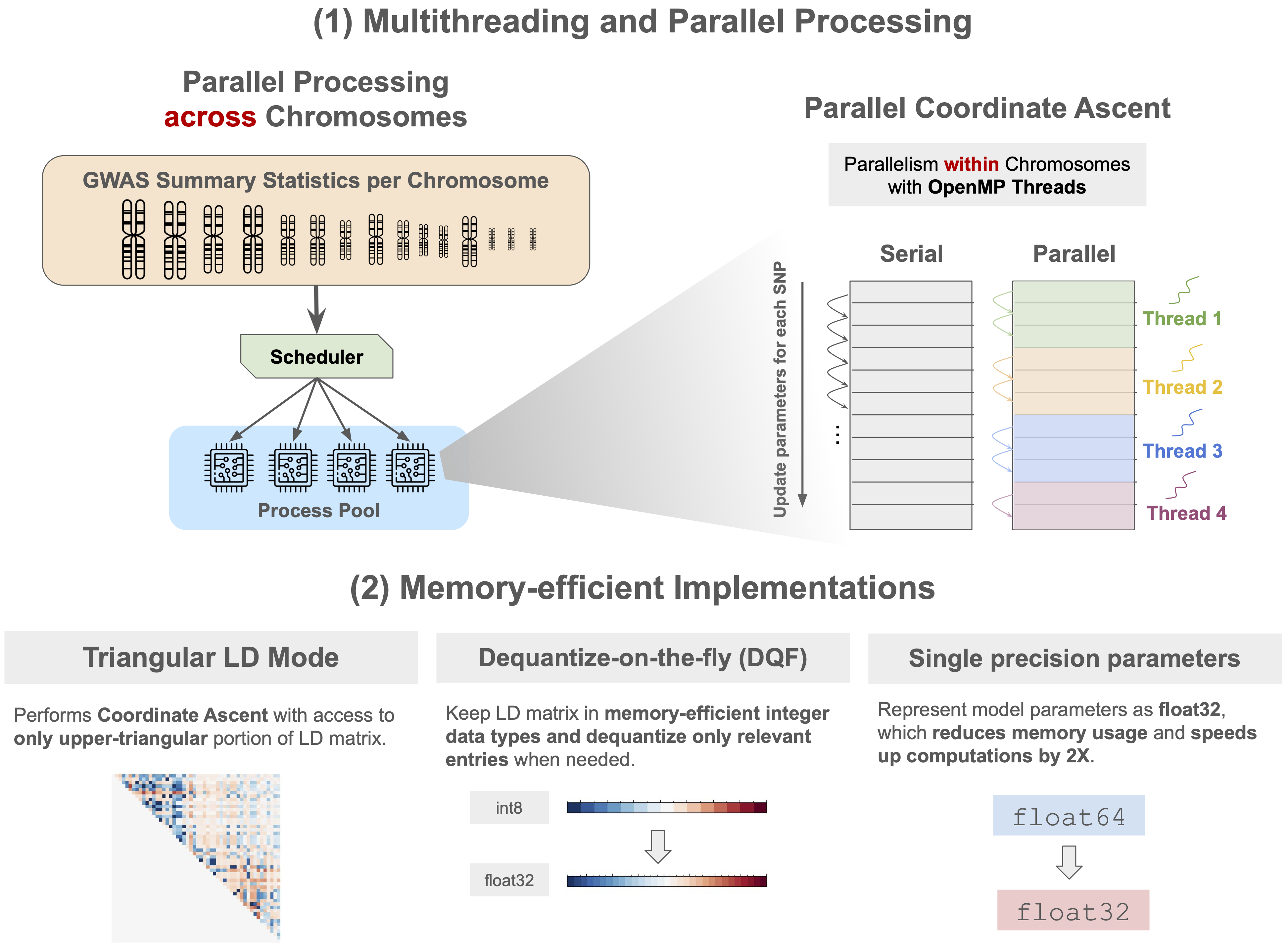
Polygenic risk score inference
Toward whole-genome inference of polygenic scores with fast and memory-efficient algo-rithms American Journal of Human Genetics 2025
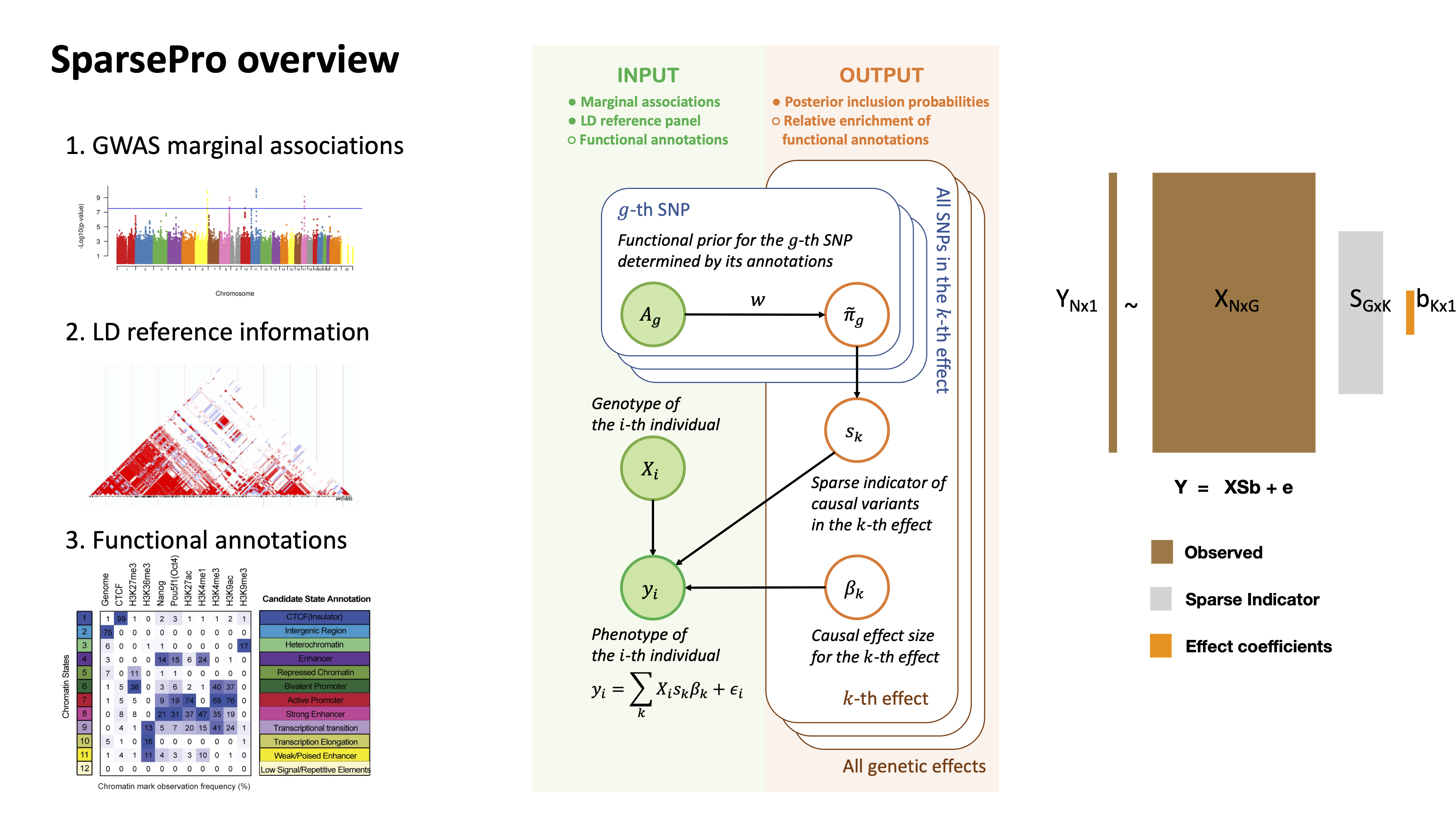
Inferring causal genetic variants
SparsePro: an efficient genome-wide fine-mapping method integrating summary statistics and functional annotations. PLOS Genetics 2023
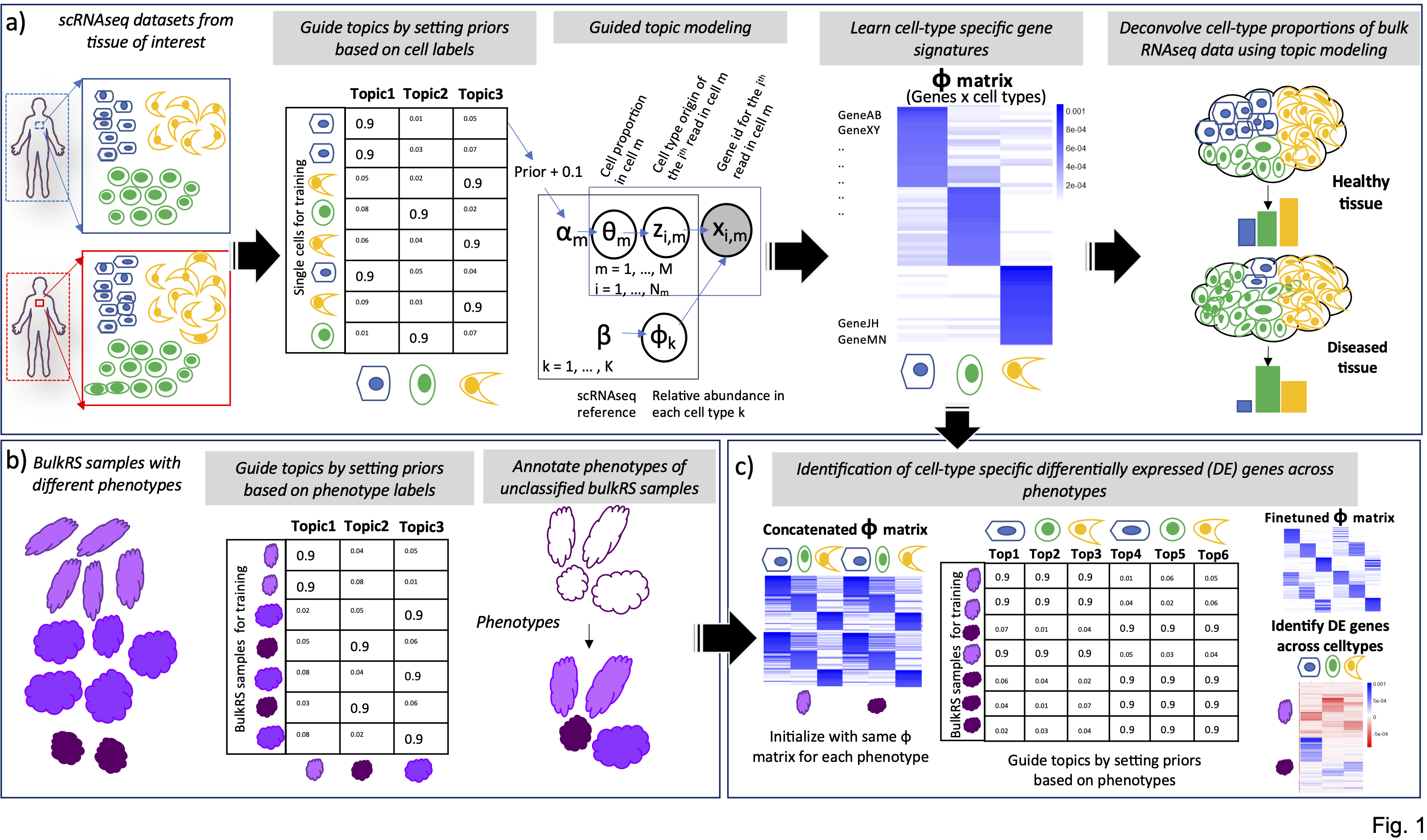
Cell-type deconvolution
Guided-topic modelling of single-cell transcriptomes enables joint cell-type-specific and disease-subtype deconvolution of bulk transcriptomes with a focus on cancer studies. Genome Biology 2023
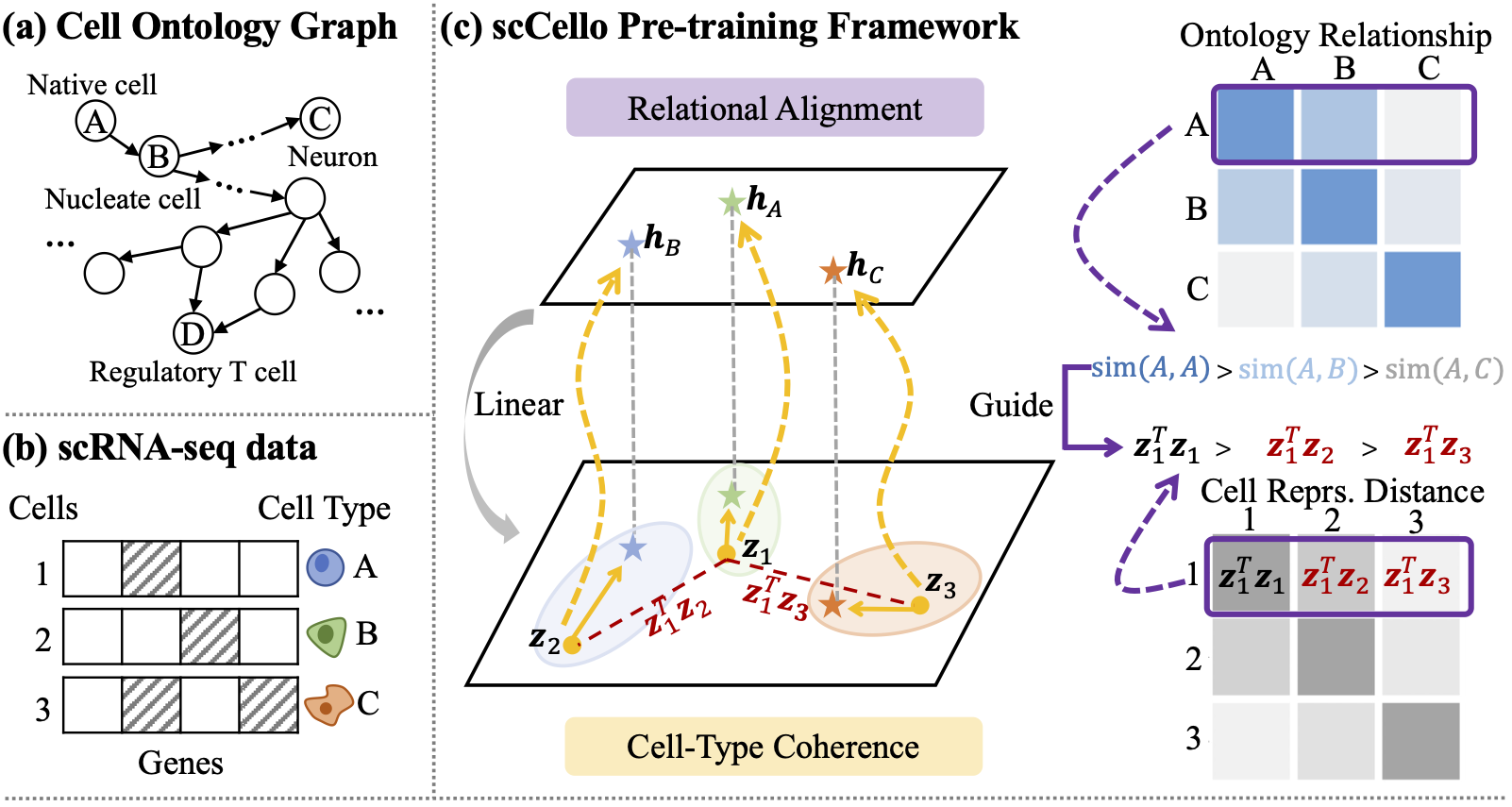
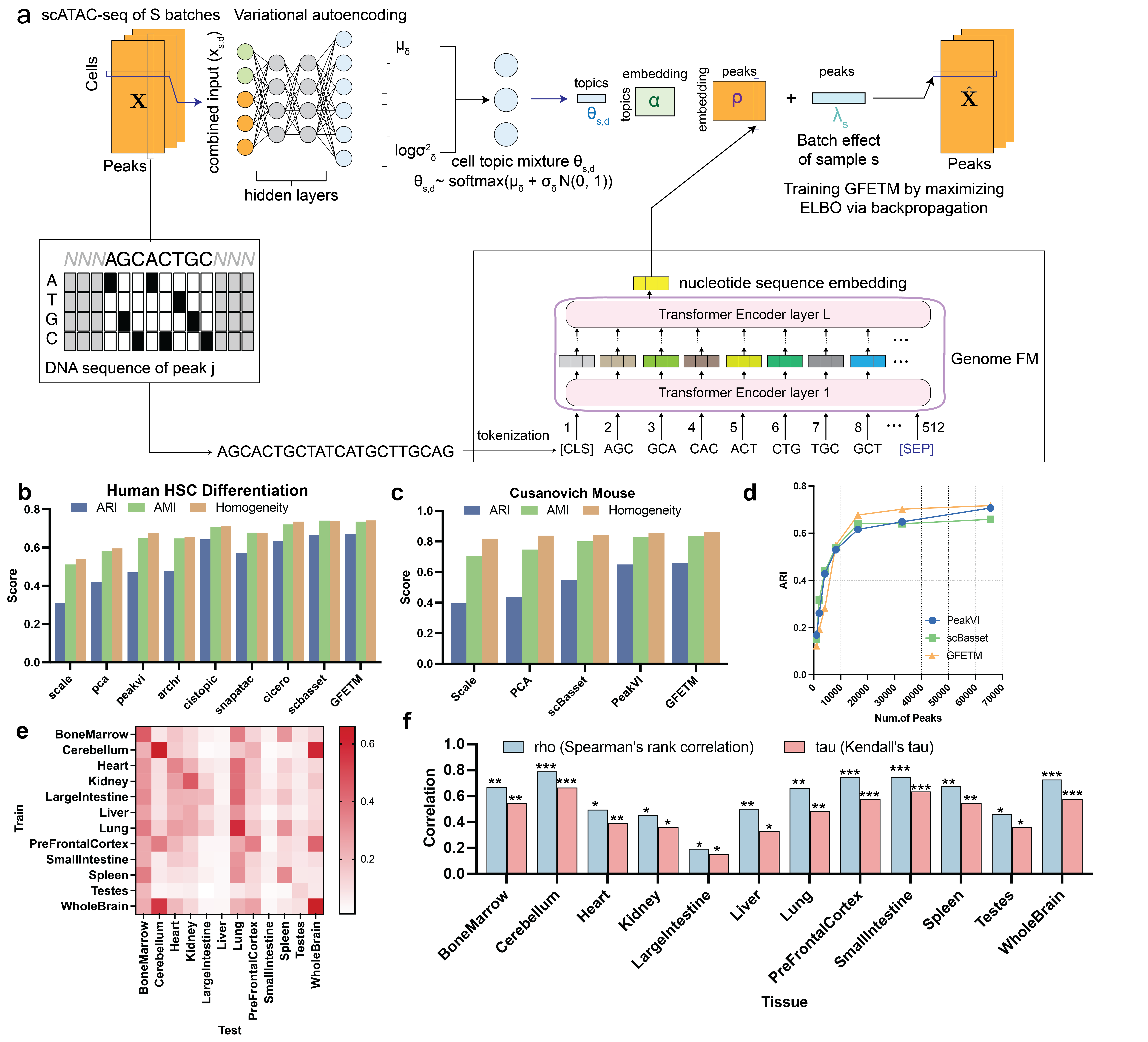
Single-cell foundation model
Cell ontology guided transcriptome foundation model. NeurIPS 2024 spotlight.