Table Of Contents
Previous topic
Next topic
Quick search
Enter search terms or a module, class or function name.
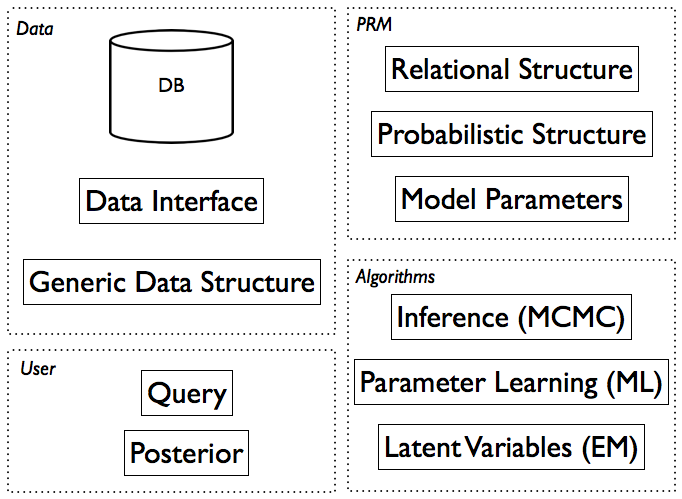
ProbReM framework structure
The illustration above shows the structure between the ProbReM packages and modules. The Modelling Workflow section below is a walk through for designing a new model we call PRMexample. Please refer to the tutorial in the Example Model for details.
A ProbReM project is a python script used to specify a PRM, connect it to database and configure the algorithms that will be used. The last section describes the structure of such a script.
The data itself is stored in a separate relational database, ProbReM accesses the data using a data.datainterface which makes the choice of database irrelevant (theoretically). Currently ProbReM supports the SQLite format which is implemented in data.sqliteinterface.SQLiteDI.
Assuming the data is stored in ./data/database.sqlite, a data interface specification is defined in XML and saved in the file ./DIexample.xml:
<?xml version="1.0" ?>
<DataInterface name="DIexample">
<Crossvalidation folds='1'>
<Dataset type='SQLite' path='./data/database.sqlite'/>
</Crossvalidation>
</DataInterface>
Which is a simple example where just one data source is specified. It is also possible test the model using cross validation by specifying multiple data sources, in which case the data has to be split up on the database level. Otherwise the different folds would have to be accessed by querying one database which decreases the performance. The XML parser for the data interface xml_prm.parser.DataInterfaceParser contains the specifications for the tags.
The Ground Bayesian Network (GBN) is a generic data structure (a graph) that contains the data necessary to answer a given query. The GBN is stored in propositional form, as opposed to the first-order representation of the PRM, thus only the subgraph which d-separates the full graph given the query is loaded. The network.groundBN module implements this data structure. The inference engine is loading the GBN using the method inference.engine.unrollGBN().
The PRM model is also specified in XML and saved in the file, e.g. ./PRMexample.xml:
<?xml version="1.0" ?>
<PRM name="PRMexample" datainterface="./DIexample.xml" >
<RelationalSchema>
<Entities>
<Entity name="A">
<Attribute name="Aa" type="Binary"/>
</Entity>
<Entity name="B">
<Attribute name="Ba" type="Integer" description="1,20"/>
</Entity>
[......]
</Entities>
<Relationships>
<Relationship name="AB" foreign="A.pk,B.pk" type="1:n">
<Attribute name="ABa" type="Binary"/>
</Relationship>
[......]
</Relationships>
</RelationalSchema>
<DependencyStructure>
<Dependency name="Aa_Ba" child="A.Aa" parent="B.Ba" constraints="A.pk=B.pk" aggregator='AVG'/>
[......]
</DependencyStructure>
<LocalDistributions>
<LocalDistribution attribute='A.Aa' file='./localdistributions/Da_Aa.xml'/>
<LocalDistribution attribute='B.Ba' file='./localdistributions/Ba_Aa.xml'/>
<LocalDistribution attribute='AB.ABa' file='./localdistributions/Ca_Aa.xml'/>
</LocalDistributions>
</PRM>
A list of all possible tags as well as attributes, please see the documentation of the XML parser xml_prm.parser.PRMparser used by ProbReM. The PRM is defined by the relational structure <RelationalSchema>, probabilistic structure (<DependencyStructure>) and the model parameters (<LocalDistributions>; the conditional probability distributions of the attributes).
Model Parameters:
Usually the local conditional probability distributions (CPDs) are learned from data. ProbReM uses a Maximum Likelihood (ML) estimate which is implemented in learners.cpdlearners.CPDTabularLearner. Currently only tabular CPDs are supported, prm.localdistribution.CPDTabular. The learner instance can be configured to save the distributions to a file, the necessary files will be created automatically and the parser loads the distributions from disk if a corresponding file is available.
Inference:
All inference methods in ProbReM are based on Markov Chain Monte Carlo methods. MCMC has proven to be very efficient, unfortunately MCMC algorithms in practice require a lot of fiddling around with parameters, e.g. burn in, number of samples collected, proposal distribution, convergence diagnostics. Depending on the type of query, different algorithms with different parameters are necessary. For this reason a ProbReM project has to allow a flexible configuration of the inference method. More complex models may also require custom proposal distributions. The inference module offers the framework for MCMC inference, please refer to the documentation for details.
A python script is used to configure a ProbReM project:
probremI = Probrem()
''' PRM '''
prmSpec = "PRMexample.xml"
probremI.prmI = config.loadPRM(prmSpec)
''' DATA INTERFACE '''
diSpec = probremI.prmI.datainterface
#diSpec = "DIexample.xml"
probremI.diI = config.loadDI(diSpec)
#configure datainterface with the prm instance
probremI.diI.configure(probremI.prmI)
Next, the local distributions are learned from data. If the probabilistic structure and the data don’t change, the CPDs can be loaded from disk the next time the PRMexample model is instantiated.
''' LEARNERS '''
#we load a cpd learner to learn the CPDs for our attributes
probremI.learnersI['ourCPDlearner'] = config.loadLearner('CPDTabularLearner')
#we configure the learner to use the prm and data interface we instantiated
probremI.learnersI['ourCPDlearner'].configure(probremI.prmI,probremI.diI,learnCPDs=False)
probremI.learnersI['ourCPDlearner'].learnCPDsFull(saveDistributions=True,forceLearning=True)
After the model parameters are defined the inference method can be configured. The parameters are needed as engine precomputes as much as possible, e.g. the likelihood functions in the case of a Gibbs sampler.
''' INFERENCE ENGINE '''
probremI.inferenceI = config.loadInference('MCMC')
#we configure the engine to use the prm and data interface we instantiated
probremI.inferenceI.configure(probremI.prmI,probremI.diI)
Assuming that the script is saved in ./probremExample.py, the model can now be used for queries by creating another script ./queryExample.py which imports the model specification. A very simplistic example is given below:
from probremExample import *
# creating a query
exQuery = Query(event,evidence)
probremI.inferenceI.infer(exQuery)
# display the cumulative mean to test the convergence
posterior.cumulativeMean()
The Example Model includes more details as well as complete source code.